Examples¶
These examples demonstrate some of the basic use-cases of dicomtools.
Reading DICOM Data¶
You can either read a single DICOM file, multiple DICOM files from the same series, or a DICOMDIR file. A dicomtools.series.DicomSeries object will be returned, except in the case of dicomtools.dicom_read.read_dicomdir(), which will return a list of dicomtools.series.DicomSeries objects.
Reading a DICOMDIR file:
>>> import dicomtools
>>> dicomtools.dicom_read.read_dicomdir('data/DICOMDIR')
[<dicomtools.series.DicomSeries object at 0x00000000054A9EB8>,
<dicomtools.series.DicomSeries object at 0x0000000007EF3710>,
<dicomtools.series.DicomSeries object at 0x0000000007EF3DD8>]
Reading series of DICOM files:
>>> import dicomtools
>>> dicomtools.read_dicom_series(['data/im_001.dcm', 'data/im_002.dcm', 'data/im_003.dcm'])
<dicomtools.series.DicomSeries object at 0x0000000008EF85C0>
Reading a single DICOM file:
>>> import dicomtools
>>> dicomtools.read_dicom('data/im_dat.dcm')
<dicomtools.series.DicomSeries object at 0x0000000006718128>
Building a 3D Volume Dataset¶
If a dicomtools.series.DicomSeries object represents 3D data as a multi-frame dataset or multiple DICOM files within a series, you can easily process this using the dicomtools.volume.DicomVolume.
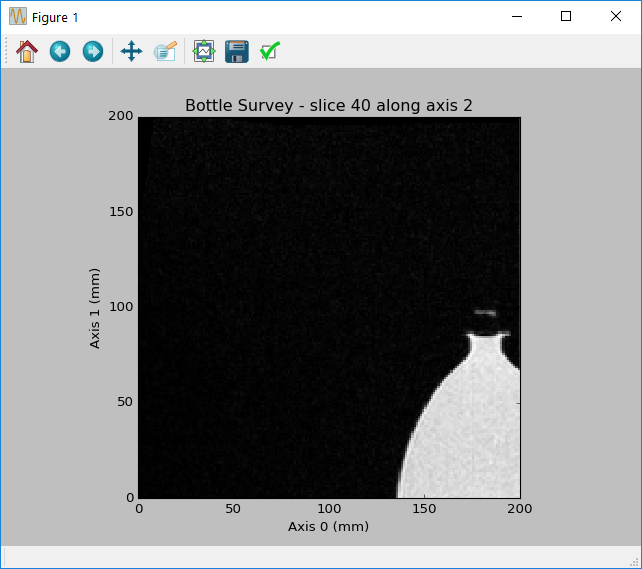
View a single slice of the 3D multi-frame dataset:
>>> import dicomtools
>>> series = dicomtools.read_dicom('data/im_dat.dcm')
>>> volume = dicomtools.volume.DicomVolume(series)
>>> dicomtools.visualization.plot_slice(volume, 2, 40)
Moving Between Image/Pixel and DICOM Patient Coordinates¶
The dicomtools.coordinates module contains useful functions for transforming points between
coordinate systems, but with a dicomtools.volume.DicomVolume object, it’s much simpler.
Transform pixel (5,5) to the position in patient coordinates:
>>> volume = dicomtools.volume.DicomVolume(series)
>>> img2pat = volume.build_image_to_patient_matrix()
>>> img2pat
array([[ 0. , 0. , -1.2 , 59.4 ],
[ 1.136, 0. , 0. , -180. ],
[ 0. , -1.136, 0. , 180. ],
[ 0. , 0. , 0. , 1. ]])
>>> pixel_position = [5, 5]
>>> dicomtools.coordinates.transform_vectors(img2pat, pixel_position)
array([ 59.4 , -174.318, 174.318])
Transform pixel (4,5,6) to the position in patient coordinates:
>>> volume = dicomtools.volume.DicomVolume(series)
>>> img2pat = volume.build_image_to_patient_matrix()
>>> img2pat
array([[ 0. , 0. , -1.2 , 59.4 ],
[ 1.136, 0. , 0. , -180. ],
[ 0. , -1.136, 0. , 180. ],
[ 0. , 0. , 0. , 1. ]])
>>> pixel_position = [4, 5, 6]
>>> dicomtools.coordinates.transform_vectors(img2pat, pixel_position)
array([ 52.2 , -175.455, 174.318])
Transform a position in patient coordinates back to a pixel position:
>>> import numpy as np
>>> volume = dicomtools.volume.DicomVolume(series)
>>> img2pat = volume.build_image_to_patient_matrix()
>>> pat2img = np.linalg.inv(img2pat)
>>> pat2img
array([[ 0. , 0.88 , 0. , 158.4 ],
[ -0. , -0. , -0.88 , 158.4 ],
[ -0.833, -0. , -0. , 49.5 ],
[ 0. , 0. , 0. , 1. ]])
>>> dicomtools.coordinates.transform_vectors(pat2img, [59.4, -174.318, 174.318])
array([ 5., 5., 0.])
>>> dicomtools.coordinates.transform_vectors(pat2img, [52.2, -175.455, 174.318])
array([ 4., 5., 6.])
Transform multiple positions simultaneously (faster than transforming each individually):
>>> volume = dicomtools.volume.DicomVolume(series)
>>> img2pat = volume.build_image_to_patient_matrix()
>>> img2pat
array([[ 0. , 0. , -1.2 , 59.4 ],
[ 1.136, 0. , 0. , -180. ],
[ 0. , -1.136, 0. , 180. ],
[ 0. , 0. , 0. , 1. ]])
>>> pixel_positions = [[4,5,6], [5,5,0]]
>>> dicomtools.coordinates.transform_vectors(img2pat, pixel_positions)
array([[ 52.2 , -175.455, 174.318],
[ 59.4 , -174.318, 174.318]])
Exporting DICOM Images to PNG¶
The dicomtools.export module contains useful functions for exporting images.
Exporting a dicomtools.volume.DicomVolume pixel data to multiple image files:
>>> volume = dicomtools.volume.DicomVolume(series)
>>> volume.export_images('images_dir', 'image')
Exporting a single image slice:
>>> volume = dicomtools.volume.DicomVolume(series)
>>> im_data = volume.info['pixel_data'][:,:,10]
>>> dicomtools.export.export_image_to_png(im_data, 'images_dir/image.png')